

The element Molecules of type MoleculesType is a container for one or more Molecule elements of type MoleculeType.
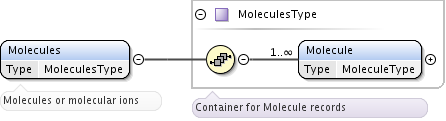
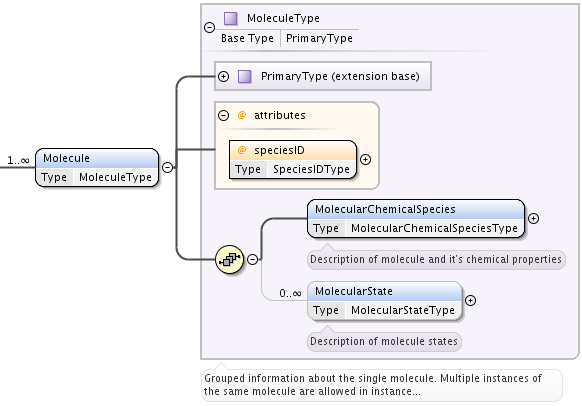
This element is defined as MoleculeType, which is an extension of PrimaryType. It provides all information on molecular chemical species through MolecularChemicalSpecies of type MolecularChemicalSpeciesType and one or more elements MolecularState of type MolecularStateType. These molecular species may be involved in various processes that are described in the Processes section. Referencing is done either by speciesID or stateID attributes, or both.
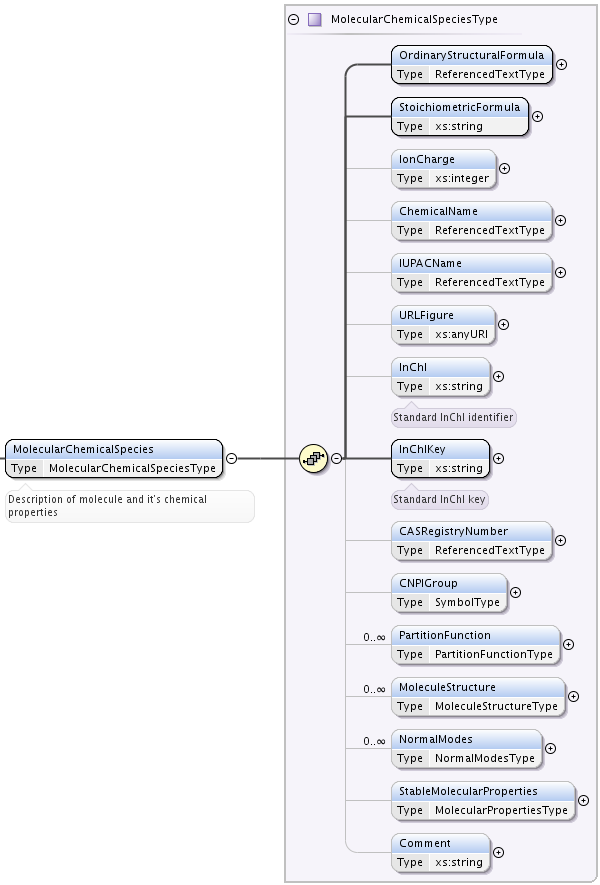
The element MolecularChemicalSpecies describes a simple model to identify the chemical molecule involved in the description of States and Processes.
NOTE: Recommendations on Organic and Biochemical Nomenclature, Symbols and Terminology are available on the International Union of Pure and Applied Chemistry [IUPAC] webpage.
MolecularChemicalSpecies element has following child elements defined:
OrdinaryStructuralformula mandatory element, of type ReferencedTextType. Standard formula describing the chemical complex written in Latex (molecule or molecular ion). For the time being, the ordinaryStructuralformula element can not be used for search, because a worldwide consensus among molecular physicists is not reached yet. Here are some guidelines on how to fill this element, based on kind of molecule:
For simple molecules involving several atoms, the formula should reflect the order of the chemical bonds involved.
Isotopic atomic species should be described by the usual chemical element name (see List of Atomic Elements) with the atomicMassNumber as an upper left subscript. For example: Carbon 13 would be $^{13}C$ (
).
For the special case of Hydrogen, isotopic atomic species have specific symbols and names: Symbol= D and Name= Deuterium for core with 1 neutron and 1 proton, Symbol= T and Name= Tritium for core with 2 neutrons and 1 proton.
When one or several radicals, such as methyl,
, are involved, they should be indicated in bracket followed by the number of their occurence as a lower subscript. They are placed in the formula following the order of the chemical bonds. Sometimes the formula of the radical is replaced by an alias such as (Me) for methyl or (Et) for ethyl.
The conformation (cis, trans, etc ...) should be indicated before the part of the formula giving the various atomic components.
For more complex molecules, no general rules are provided for now in the document and any string describing the molecule can be used.
Molecular ions should be described by their chemical formula followed by a plus/minus sign and the number of charges (when different from one). For example, the dyazenylium molecular ion would be $N_2H^+$ (
) and the carbonyl doubly charged ion $CO^{2+}$ (
).
StoichiometricFormula mandatory string element. For molecules it is constituted by an alphabetical suite of the atomic constituents followed by the total number of their occurence (purely ASCII). For example: CH2O2 (
) corresponding to formic acid whose formula is $t/c-HCOOH$ (
). This is useful for a primary search of resources.
IonCharge optional integer element. It gives the charge of the molecular ion. Examples: +1 or -1
ChemicalName element of type ReferencedTextType, a string (with reference) giving the name of the chemical complex. The ChemicalName element can not be used for search, but rather for information because different names might be associated to a single chemical complex.
IUPACName element, of type ReferencedTextType, a string (with reference) giving the IUPAC name. It can be used for search.
URLFigure optional element, that provides an URL to a figure showing the molecule in its stable configuration.
InChI optional string element. Provides the InChI identifier. [InChI]
InChIKey mandatory string element. Provides the InChIKey hash of InChI identifier. This element is mandatory because InChIKeys appeared to be the most convinient and relatively reliable way to identify if two Molecule blocks of data, originating from different databases, are describing the same species or not.
CASRegistryNumber optional element of type ReferencedTextType that provides the CAS Registry Number.
CNPIGroup element of type SymbolType:, that describes the Complete Nuclear Permutation Inversion Group.
PartitionFunction element that may contain a temperature dependence of molecule partition function in a form of a list of points. May be specified multiple times.
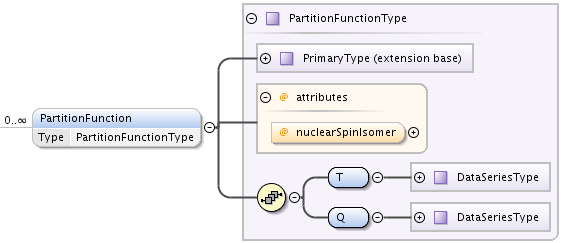
Extending PrimaryType, it has:
- nuclearSpinIsomer optional attribute to define nuclear spin isomer, the same way as in MolecularStateCharacterisation.
- T element of type DataSeriesType to define temperature points
- Q element of type DataSeriesType to define partition function values
MoleculeStructure optional element that is a link to [CML] description of molecular structure. Extending PrimaryType, it defines additionally
- optional electronicStateRef attribute of type StateRefType that may give a link to a separate description of electronic state,
- mandatory atomArray element from [CML], that must occur at least once,
- optional bondArray element from [CML], that may occur multiple times.
This description should be sufficient to define the structure of any complex molecule.
NormalModes element for description of vibrational normal modes of complex molecules.
StableMolecularProperties element if type MolecularPropertiesType which provides information on properties of the molecule.
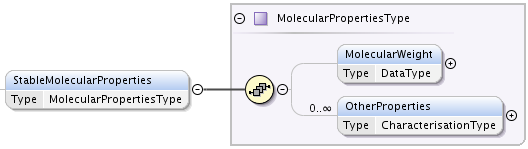
MolecularPropertiesType has two elements:
- MolecularWeight, of type DataType, that carries the sum of the individual isotopic masses of all the atoms in a molecule,
- multiple OtherProperties element of type CharacterisationType that allows to specify arbitrary properties of molecule in a form of a named dataset.
Comment string element for arbitrary comments.
Formally a MolecularState element of type MolecularStateType is characterized by a single eigenvalue (possibly degenerate) and a single eigenstate (when non degenerate eigenvalue) of the hamiltonian describing the energy structure of the chemical compound. When the eigenvalue is degenerate, the quantum numbers associated to the degeneracy are not provided.
The eigenvalue corresponds to the StateEnergy, and is given relative to an energyOrigin. The eigenstate is characterized by a set of good quantum numbers, such as parity and total angular momemtum, and described by a wavefunction often expanded over some basis functions. The expansion is characterized by a coupling scheme between the quantum numbers identifying individual basis functions.
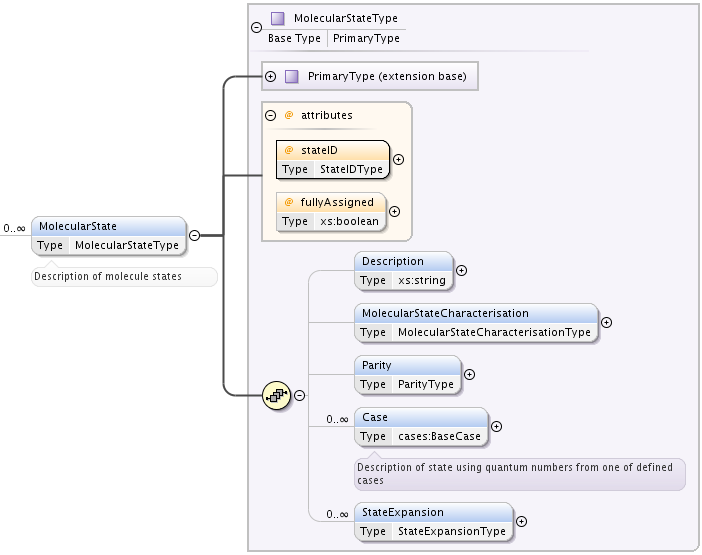
Following elements and attributes are defined for MolecularState
- stateID mandatory attribute, of type StateIDType, used for referencing the particular state.
- fullyAssigned optional boolean attribute, “true” defines that the state is fully described by a set of good quantum numbers.
- optional Description string element, where arbitrary name of state can be given.
- optional MolecularStateCharacterisation element of type MolecularStateCharacterisationType that describes all quantities related to the molecular state apart from quantum numbers (e.g. statistical weights, Land’e factors, radiative lifetime of the level and other properties).
- optional Parity element, that gives the total parity of the level. It is of type ParityType, a token taking the values odd or even.
- optional Case element of type BaseCase, that is a container for [case-by-case] definition of quantum numbers. BaseCase is explained below.
- optional StateExpansion element that allows to describe the state as a superposition of basis quantum numbers sets, each set with it’s own coefficient.
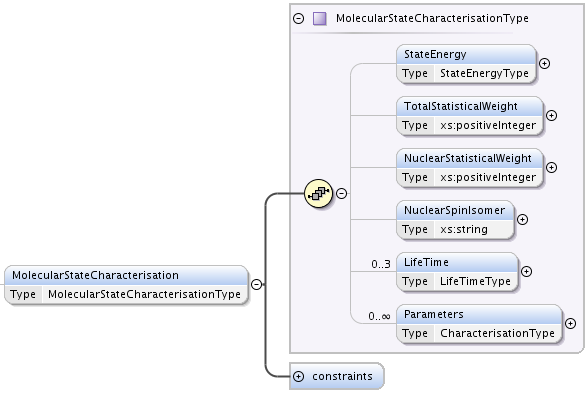
MolecularStateCharacterisation element, defined as MolecularStateCharacterisationType, describes all quantities related to the molecular state apart from it’s quantum numbers. Following optional elements are defined:
StateEenrgy, of type StateEnergyType. Defined as an extension of DataType with an additional optional string attribute energyOrigin. energyOrigin should contain a human readable string indicating the nature of the origin taken for the energy scale. For example, energyOrigin might be ground electronic, vibrational, rotational state or could be ionization energy or dissociation energy. By physical meaning, state energy is the eigenvalue of the hamiltonian describing the species.
TotalStatisticalWeight positive integer element, keeps statistical weight associated to the level, with all degeneracies (including nuclear spins).
NuclearStatisticalWeight positive integer element, the same as TotalStatisticalWeight but for nuclear spins only.
NuclearSpinIsomer, a string indicating the type of nuclear spin symmetry. Possible values can be para, ortho, meta, A, E. This element is a comfort element very often used to classify levels.
Lifetime, of type LifeTimeType. Defined as an extension of DataType with an additional mandatory attribute decay that may take values total, totalRadiative, totalNonRadiative, allowing to represent total state lifetime, including radiative and non-radiative decay mechanisms, or define them separately.
Maximum of three occurences of this element is possible, each must have different value in decay attribute.
When only discrete radiative decay is involved, it is given by
Parameters, list of elements of CharacterisationType. It allows to add any additional characterisation of the molecular state.
If data producer is intending to use this element, full description of data format and meaning should be given in one of the source references.
As an example it can be used to describe a statistical weight associated to the level including some degeneracies, but not all. In that case the element “Parameter.Name” takes the value PseudoStatisticalWeight. It does not include all degeneracies and is used in fractions. CDMS database provides such PseudoStatisticalWeight.
The case-by-case XML description of molecular states within VAMDC-XSAMS is designed to provide a straightforward and flat data structure for representing the quantum numbers and symmetries that denote a molecular state. The reader is the refered to the [case-by-case] documentation for full description of the various cases.
Each MolecularState has Case element of type cases:BaseCase from separate namespace.
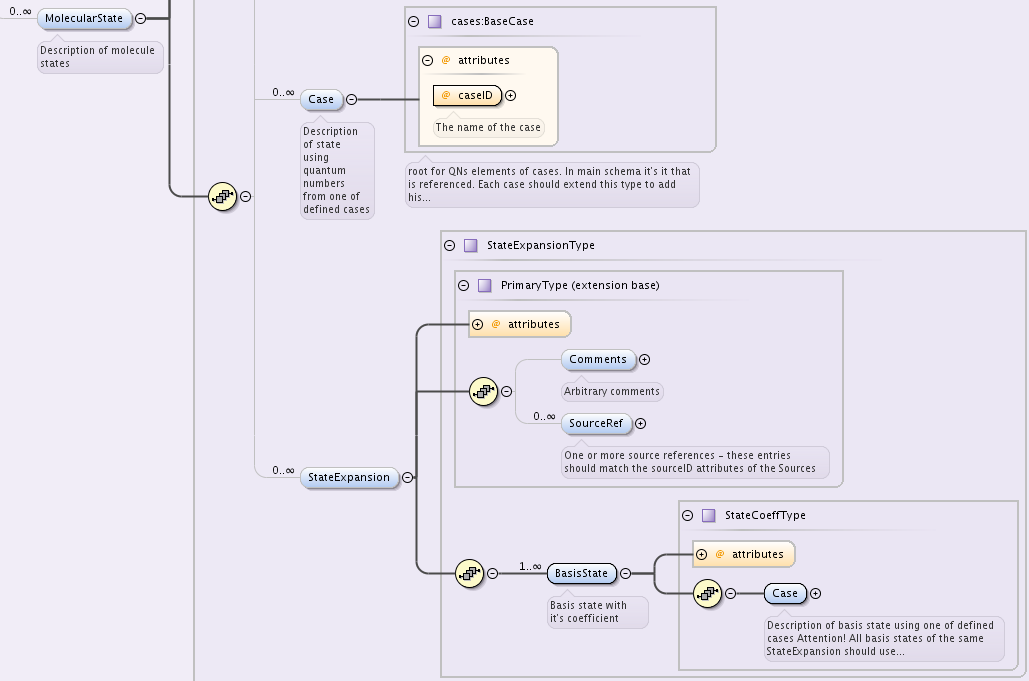
BaseCase type defines the single attribute, caseID, that denotes the case used.
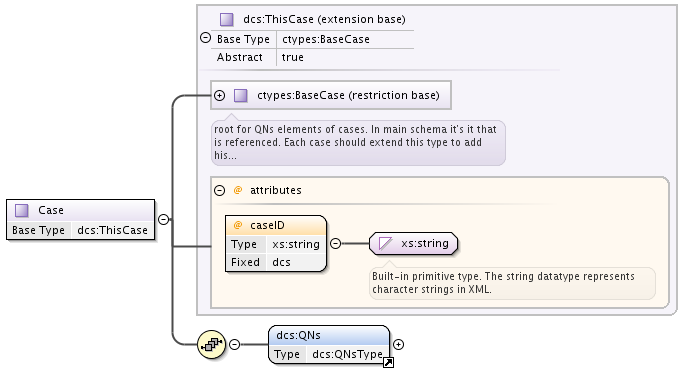
Each specific case, in turn, extends that BaseCase type, defining specific caseID attribute value and adding QNs element that contains a sequence of quantum numbers and symmetries.
Here, specific XML types, used only in Species.Molecules are described.
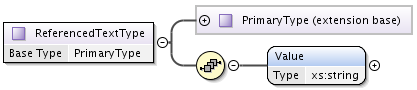
An extension of PrimaryType that has additional string Value element, is used to define strings with Source reference.
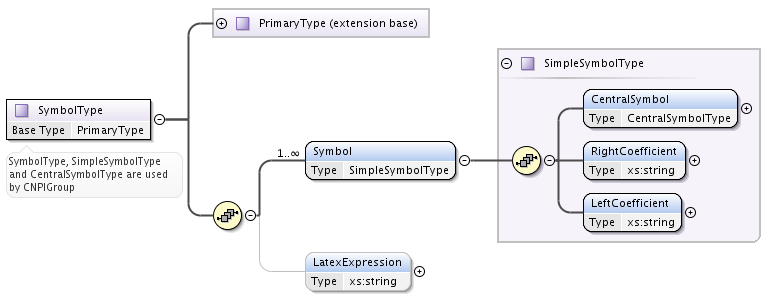
It is a model to describe symbols. Extending PrimaryType, it adds:
- a list of Symbol elements of type SimpleSymbolType
- a string element LatexExpression, that may contain latex representation of the symbol.
SimpleSymbolType defines three string elements,
CentralSymbol which is a string with 4 string attributes,
- UpperLeftValue
- LowerLeftValue
- UpperRightValue
- LowerTightValue
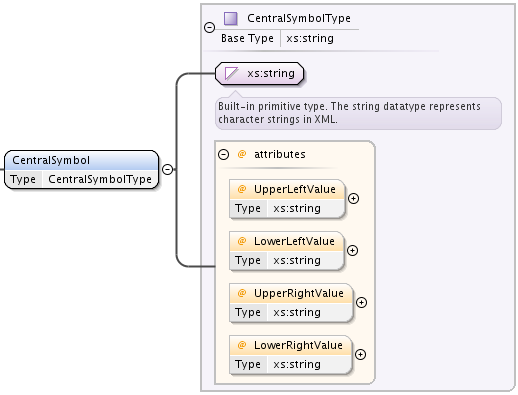
RightCoefficient
LeftCoefficient
Mathematical symbols and greek alphabet letters should be defined as unicode symbols, for example, ∞ or ∞ for infinity sign (
)
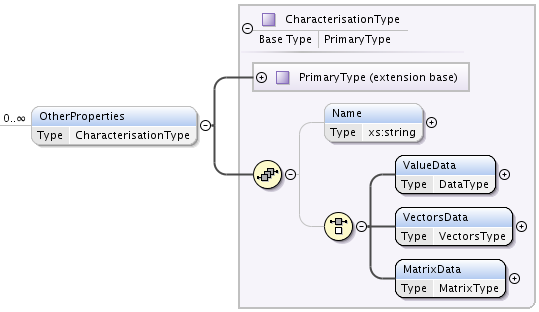
CharacterizationType is an extension of PrimaryType, adding a Name string element and a choice of one of:
- ValueData of type DataType,
- VectorsData of type VectorsType or
- MatrixData of type MatrixType:.
permitting representation of arbitrary data, relevant to state.
To represent vibrational normal modes of molecules, NormalModes element is used.
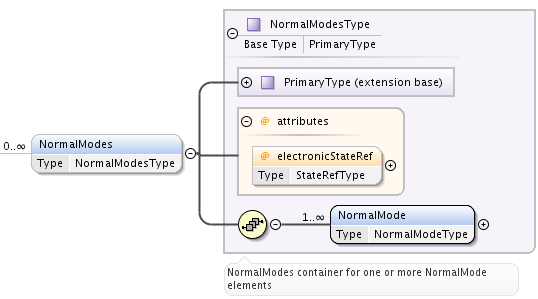
Each NormalModes element, extending PrimaryType, may have an attribute electronicStateRef, defining reference to electronic state, and must have at least one NormalMode element, each defining a single mode.
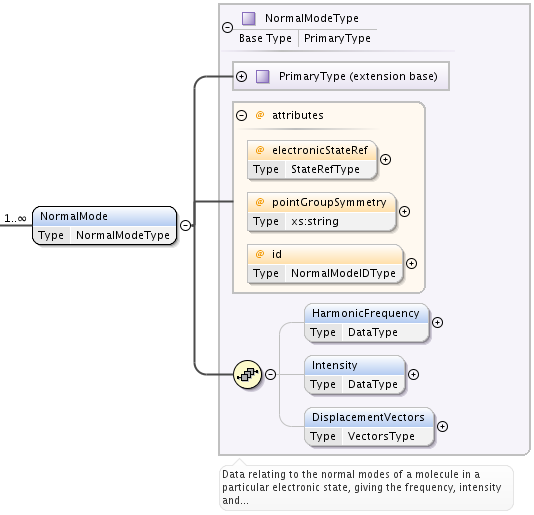
NormalMode element, also extending PrimaryType, has following attributes and elements:
- optional electronicStateRef attribute, of type StateRefType, defining electronic state;
- optional pointGroupSymmetry string attribute;
- optional id attribute of NormalModeIDType, defining unique identifier for this mode, to be referenced from radiative CrossSection band assignment;
- optional HarmonicFrequency element of DataType;
- optional Intensity DataType element;
- optional DisplacementVectors element of type VectorsType to define atoms displacement configuration of the mode.
- ref attribute of Vector must contain the id of the atom in molecule’s structure,
- x3, y3, z3 define atom’s relative displacement against it’s position in ground state.
Example XML block for NormalModes would look like:
<NormalModes electronicStateRef="SX_Azulene-1"> <NormalMode id="V1" pointGroupSymmetry="A1"> <HarmonicFrequency> <Value units="1/cm">162</Value> <Accuracy><Systematic>1</Systematic></Accuracy> </HarmonicFrequency> <Intensity> <Value units="km/mol">0</Value> </Intensity> <DisplacementVectors units="A"> <Vector ref="C1" x3="0." y3="0.001" z3="0.0005"/> <Vector ref="C2" x3="0.01" y3="-0.001" z3="0.0005"/> <Vector ref="C3" x3="-0.005" y3="0.001" z3="0."/> <!-- etc... --> </DisplacementVectors> </NormalMode> <NormalMode id="V2" pointGroupSymmetry="A2"> <HarmonicFrequency> <Value units="1/cm">214</Value> <Accuracy><Statistical>5</Statistical></Accuracy> </HarmonicFrequency> </NormalMode> <NormalMode id="V3" pointGroupSymmetry="A1"> <HarmonicFrequency> <Value units="1/cm">1720.21</Value> <Accuracy> <Statistical>0.05</Statistical> </Accuracy> </HarmonicFrequency> </NormalMode> </NormalModes>